RNA VIRUSES
Enveloped, segmented, ss, negative sense, (class V)
Orthomyxoviridae
2. Bunyaviridae
3. Arenaviridae
ORTHOMYXOVIRIDAE
General Characteristics
Small in number but large in unusual features, ranging from its epidemiology to its molecular biology and appearance.
Internal nucleocapsid and an envelope made up of an inner matrix protein, a lipid bilayer, and external glycoproteins.
The shape of the virion is not uniform: besides more or less round particles of about 100 nm in diameter, there occur somewhat elongated, often bent particles.
Virions are pleomorphic, often spherical or elongated or bent, 80-120 nm in diameter.
Highly pleomorhic -- filamentous forms occur.
Virions consist of a lipid-containing envelope, with large peplomers, 10-14 nm in length and 4-6 nm in diameter.
Peplomers are trimeric hemagglutin and tetrameric neurominidase protein structures.
Only in A and B virions.
These surface proteins, particularly the hemagglutinin, determine the antigenic specificity of the subtypes.
Within the envelope there are helically symmetrical capsids of different size classes, 150-130 nm in length with a loop at one end.
Virions are very sensitive to treatment with heat, lipid solvents, non-ionic detergents, formaldehyde, oxidizing agents.
The infectivity is reduced after exposure to irradiation.
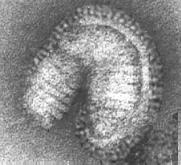
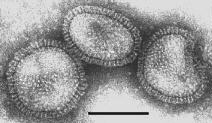
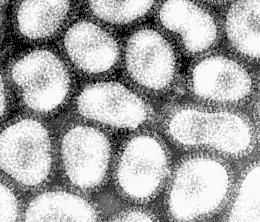
Electron micrographs of the influenza virus Type A --
Source: Linda Stannard, University of Cape Town
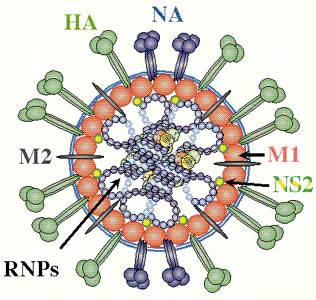
Source: Paul Digard, Dept Pathology, University of Cambridge.
Genome
- Consists of 8 (influenza viruses A and B), 7 (C and Dhori virus), or 6 (Thogoto virus) molecules.
- Linear, negative sense, ssRNA molecules.
- 10-13 kb in size.
- Segment lengths from 900 to 2,350 nucleotides.
Virions contain 7 to 9 major proteins.
Polymerase proteins (PA, PB1, PB2 in influenza A virus).
Nucleocapsid protein (NP).
Hemagglutinin (H) spikes combine with a specific carbohydrate molecule found in all eukaryotic cell membranes, the virus has the capacity to bind and clump erythrocytes.
Matrix protein (M1 or M).
Neurominidase (N) (receptor-destroying enzyme of type A that hydrolizes the protective mucous coating of the respiratory tract, assisting in viral budding and release).
Closely related viruses reassort genes during mixed infections.
Entry
- Attachment sites are probably dictated by the conformation of both spikes (N and H).
- There are fewer neuraminidase spikes (200) on the flu virion than hemagglutinating spikes (750).
- Carbohydrates in the envelope are probably responsible for the affinity of all the viruses to the surface polysaccharides of cells, including the RBC of many species, causing agglutination.
- The initial infection involves the attachment of the envelope neuraminidase spike to the cellular receptor.
- N breaks the bond holding N-acetyl-neuraminic (or sialic) acid to the end of many polysaccharide receptors on cell surfaces. Then, hemagglutination occurs.
- It appears that in certain viral strains the N also carries hemagglutinating activity.
- Entry requires an acid environment (like in Rhabdovirus).
Replication
- The first step is transcription of the (-ve sense) vRNA by the virion associated RNA-dependent RNAP to produce (predominantly) monocistronic mRNAs from each segment within the nucleus mRNAs serve as the template for subsequent genome replication.
- As with all (-) sense RNA viruses, packaging of this virus-specific transcriptase/replicase within the virus nucleocapsid is essential because no host cell contains any enzyme capable of decoding and copying the RNA genome.
- RNA segments in a mixed infection readily assort to form genetically stable hybrids within a virus.
- A mechanism must also exist that places a complete set of RNA segments inside each virion particle.
- Remarkably some proteins synthesized in the cytoplasm migrate from their site of synthesis in the cytoplasm to the nucleus.
- Since passage of molecules across the nuclear membrane is a highly selective process, this constitutes a level of control which is unique to eukaryotic cells.
- The newly formed nucleocapsid complexes coated by the M protein then enter the membrane and acquire their lipid envelope traversed by the viral spike proteins.
- The selective incorporation of the negative-sense genomic segments into the nucleocapsid most likely depends on cellular proteins.
- Subsequent budding from the cell surface occurs.
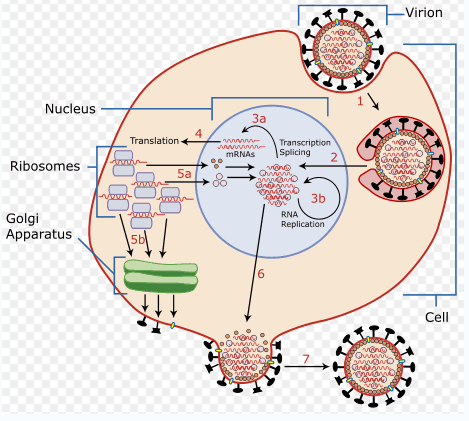
Source: w:Image:Virusreplication.png
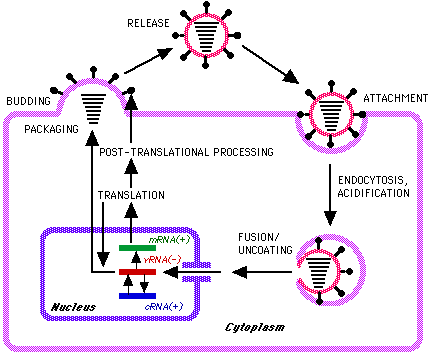
Source: Alan Cann, microbiologybytes.com
Orthomyxovirus replication takes about 6 hours and kills the host cell.
Influenza Viruses
- Types A, B and C.
- Type A is the most common with an extremely wide host range (human, pig, bird, horse). Serotypes or subtypes of Type A: H1N1, H1N2, H2N2, H3N1, H3N2, H3N8, H5N1, H5N2, H5N3, H5N8, H5N9, H7N1, H7N2, H7N3, H7N4, H7N7, H9N2, H10N7.
- Responsible for many pandemics.
- Overall mortality rate for Type A is about 0.1% of cases.
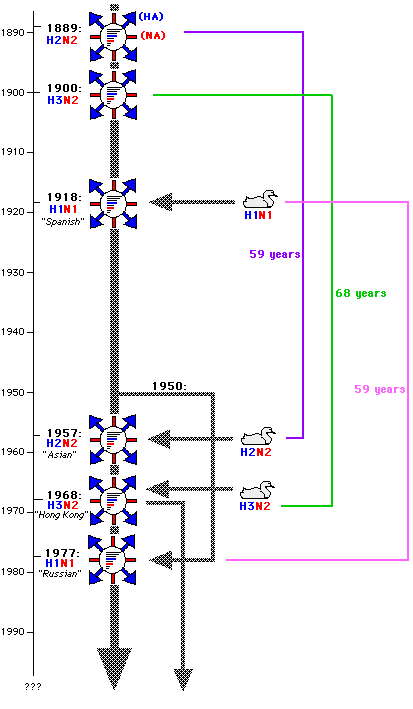
- Since 1510, 31 pandemics have been reported.
- Pandemics (1900, 1918, 1946, 1957, 1968, 1977).
- Near a million deaths in USA in 1918.
- Record: 678 fatalities in a single day in Oct 1918 in USA.
A skip-rope song:
I had a little birdie his name was enza I opened the window and in-flu-enza- Types B infect humans and seals while C infects humans and pigs. B and C do not cause pandemics
- Outbreaks of B occur every 2 or 3 years.
- Type C causes subclinical infections or mild cold-like illness.
- All three types are similar in structure and mode of replication, but their capsid proteins are different.
- Each type (A, B and C) are known for its antigenic variation or extreme variability.
- Influenza C virus has 7 RNA segments and a receptor destroying activity (a sialic acid - O- acetyl esterase) which is on the hemagglutinin protein.
- These viruses undergo changes at frequent intervals, creating new strains with different and unexpected immunological properties, is now believed to result from the accidental reassortment of RNA components of different virus strains that infect the same cells.
- Theoretically, recombination could produce 256 (28) different genome combinations.
==> A virus from a pig and a human combine and results in a virus that could spread among various hosts.
Nomenclature
It is based on the variant of antigen present.
Example: A(H3N2) that predominated in the US in 1999.
Also, the virus A(H5N1) in Asia.
Epidemiological Factors Accounting for Genetic Variation
- Degree of community resistance.
- Antigenic shift and drift.
- Animal co-infections.
- Success of mutant strain due to natural selection.
Antigenic Variation
A process of constant changes in antigenic expression in which chemical changes occurs periodically in hemagglutinin and neuraminidase, thereby yielding new strains of virus.
Types of antigenic variation:
Antigenic Drift
- Involves minor or point spontaneous mutations
that alter hemagglutinin structure.
- They accumulate and gradually give rise
to new strains with slight differences in antigenicity.
- The development of a new strain through
antigenic drift may require about 10 years.
Antigenic Shift
- Involves a major change.
- Represents the abrupt appearance of
new sero-types due to the reassortment of RNA components in cells
infected by two different strains.
- Sudden emergence of a new viral strain
due to recombination of the genome segments from different
viruses.
- Leads to epidemics.
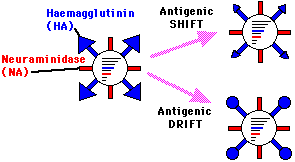
Source: Alan Cann, microbiologybytes.com
Antigenic variation precludes the development of a universally effective vaccine.
Some have been developed for particular strains.
Treatment
Active cases of influenza may be treated with amantadine (Symmetrel), and Rimantadine, drugs believed to interfere with uncoating in the replication cycle.
- Hoffman-La Roche's Tamiflu (oseltamivir phosphate, GS4104) is an inhibitor of neuraminidase taken in pill form which has been shown to confer decreased severity and duration of symptoms in clinical trials.
- Glaxo Wellcome's Relenza (Zanamivir) must be inhaled.
Vaccines
Isolated HA gives good serological protection (estimated at 60-80%).
Vaccines are produced by reassortment of egg-adapted strains with strains with the required HA type.
Large amounts of virus are then grown in embryonated eggs (cheap and efficient), purified and formalin inactivated.
The vaccine is given sub-cutaneously.
May have side effects (chills and fever).
For complete protection, it is necessary to immunize against all the different strains occurring in a given year!
Complications (major sequelas associated with flu or its vaccines).
Guillain-Barre syndrome (a neurological complication with paralysis).
Reye's syndrome (fatty degeneration of brain, liver and kidney, and neurological abnormalities).
Aspirin is forbidden in children.
{Killer flu is coming?
In May 1997, a three year-old boy died of complications of influenza in the intensive care unit of a Hong Kong hospital. This case was the first isolation of an influenza A subtype H5N1 in a human. Subtype H5 influenza viruses can cause lethal avian influenza, a disease that can decimate flocks of domestic poultry.
Fortunately, this outbreak eventually fizzled out, but not before there had been 18 confirmed human cases and 6 deaths. Why did the outbreak go away? The H5 haemagglutinin has an HA226gln sequence - i.e. was an avian virus poorly adapted to humans. There is clear evidence that between its emergence in 1997 and 2004, H5N1 isolates have become significantly more pathogenic for mammals.
All that is required now is for the virus to acquire the ability for direct human transmission and we will be in big trouble. H5N1 influenza re-emerged in Vietnam late in 2003. Also worrying, in March 1999, H9N2 viruses were isolated from two hospitalized children in Hong Kong.
Molecular analyses indicated that the HA and NA genes of the human H9N2 isolates are avian in origin and prevalent in poultry in Hong Kong. This virus has HA226leu, i.e. is a human-adapted virus, giving it the potential for rapid (pandemic?) spread in a human population which has never seen this virus before.
Is THIS the next pandemic strain? OTOH, it is now approaching 50 years since the 1957 "Asian flu" pandemic - is THIS the virus waiting in the wings to make a comeback?
So what's the worst that could happen?- A pandemic of human-adapted avian influenza such as the 1997 H5N1
virus.
- Such a reassortant could easily have a mortality rate of 30-40%.
- Within a few months 10-25% of the world's population could have been
infected.
- 6.3 billion * 0.4 * 0.25 = over half a billion deaths.
- or worse ... }*